Compound names and The InterWeb
We have a long-standing interest in finding clinical stage compounds from the literature - and it turns out that the peer reviewed literature is pretty useless, by the time something is published and appears in print it is old news, and although reviews of particular areas or targets are useful in capturing a snapshot, they are not really useful in decision support - using data to inform future experiments and investment. So these things need to be databased and online to be of much use.
So, we have a pretty big set of clinical stage kinase inhibitors that we've gathered from a wide variety of sources - this is the subject of a paper we're currently writing up, so I won't bore you with how we got the data, well, not right now.
We've posted a couple of times before about the transition of names, or classes of names as compounds go through development and approval - a search of the ChEMBL-og will show you these. But here's something hot off the press.
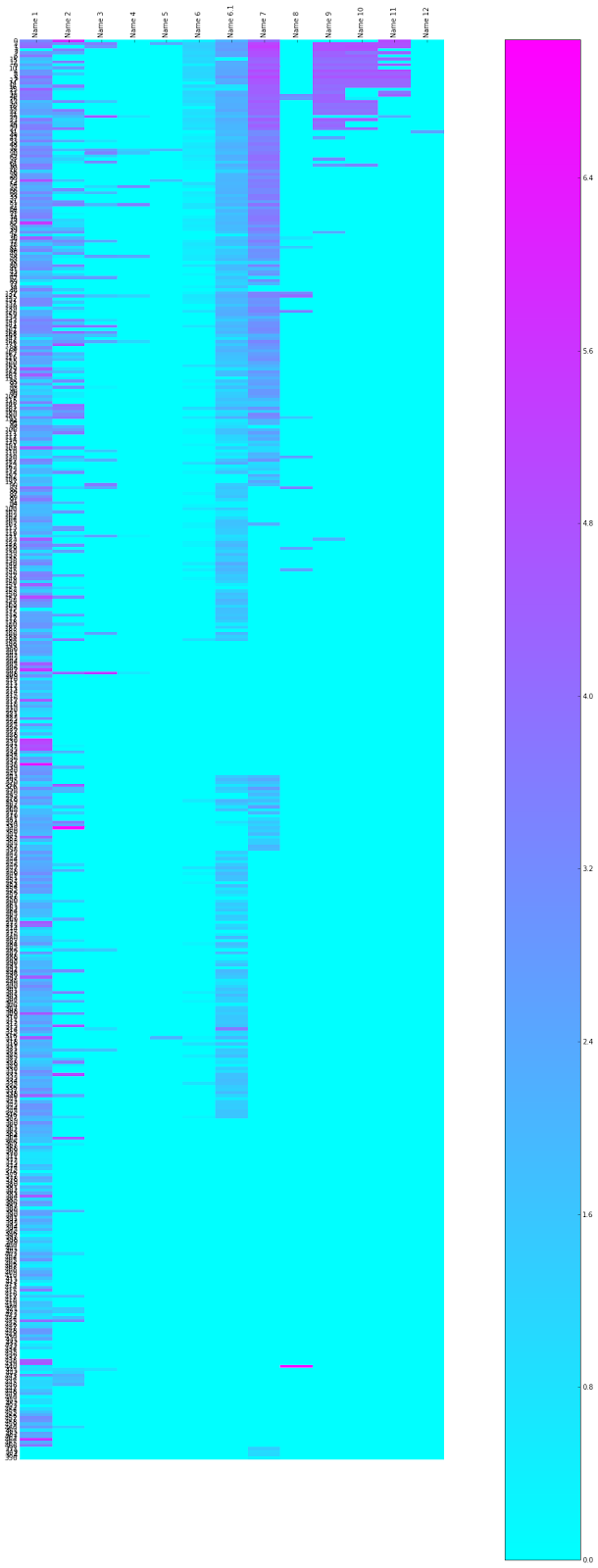
Each row is a distinct kinase inhibitor, each column is a synonym or identifier for that compound. Columns 1 to 5 are research code type names (e.g. UK-92480), with the first column being the one we use as the primary identifier, Column 6 is the InChI key, Column 6.1 is the CAS Registry number, 7 is the USAN/INN 8 is a deprecated USAN/INN or another common trivial name, Columns 9 to 12 are trade names (in case you are wondering, row 12 is of the Chinese tradename of a compound). The cells are coloured by the log(10) of the count of the number of times the name occurs in a google search - pink high, blue low (or there is no synonym of that class for that compounds). There are some false positives, where the name is unusually common, so it matches the name of something unrelated to a kinase inhibitor or therapeutic application.
It's interesting to see the diversity of names increasing as a compound becomes a launched drug, but also the broad coverage for many of these compounds with hits to the InChI key - and also for what fraction compound structures are known.
The most frequent hits in google are to:
ALL-3 11000000
CIF 7400000
X-82 1930000
AT-877 623000
X-396 425000
DE-10 362000
AC-220 245000
R-112 226000
Imatinib 226000
Erlotinib 148000
Note this is without any filtering, or other fancy stuff, just raw search engine hit counts.
Francis and jpo