-
Crimewatch?
-
UniChem - An EBI compound structure cross-referencing resource
We have faced for some time some issues with compound integration with ChEMBL - specifically the loading of compound sets into ChEMBL for cross referencing, between for example, ChEBI, PDBe compounds, etc. The ChEMBL update cycle is relatively slow with respect to some other resources, and there is inevitable thrash with compounds not being present, especially for exciting new data. Without doing something different for compound integration, we were starting to face a scenario where we had a compound table with many millions of compounds without any bioactivity data, and following this the inevitable slowdown in searching, etc.
We also had some issues facing us about curation of other people's primary data, changing compound structures, or their rendering, etc.
So, we decided to set up an external system to resolve cross-references between various databases. This is a very simple Standard InChI lookup, containing compounds from resources such as ChEMBL, ChEBI, PDBe, DrugBank, KEGG, BindingDB, PubChem, and so forth. UniChem can also handle versioning of the contained resources. We will be migrating various components of the current ChEMBL interface across to use web services on UniChem, this way, the cross links will always be fresh and correct, and we can focus on curation and optimisation of ChEMBL content. There are some other resources, like ZINC, STITCH, and ChemSpider, for example, that would be great to integrate, if we can get hold of the required data.
The easiest way for us to handle deposition into UniChem is for us to take an ftp: feed of a simple table of resource_id, standard_InChI, and standard_InChI_key.
At the moment, UniChem sits behind our firewall, but if people want to have a play, let us know.
We will write something more specific and detailed, but would welcome thoughts of whether this resolver should be externally facing, and what other resources would be good to integrate?
The image above may or may not be the UniChem logo. -
ChEMBL Widgets Update
 We have made a couple of minor updates to the ChEMBL widgets, which include:
We have made a couple of minor updates to the ChEMBL widgets, which include:
- A new widget has been created, which displays the bioactivity results shared between a ChEMBL compound and a ChEMBL target
- A new scaling parameter allows you to vary the size of the widget
- A more informative message is provided when widget has no data to display
-
Further Depositions to ChEMBL-NTD
We're delighted to announce the availability of three distinct new datasets on the ChEMBL-NTD portal, available for download, reuse, etc.These are:- Novartis-GNF Malaria Liver Stage dataset (associated with this Science publication) (Plasmodium falciparum).
- DNDi Human African Trypanosomiasis (HAT) dataset (Trypanosoma brucei)
- DNDi Chagas Dataset (Trypanosoma cruzi).
Further details of the assays and compounds are to be found on the ChEMBL-NTD portal. The data will be integrated and loaded into a future version of ChEMBL, as well as the direct data download links. Once more, we thanks the depositors, DNDi and Novartis-GNF, for their benevolence and commitment to Open Science.The associated publication for the Novartis-GNF dataset is:
%T Imaging of Plasmodium Liver Stages to Drive Next-Generation Antimalarial Drug Discovery %A S. Meister %A D.M. Plouffe %A K.L. Kuhen %A G.M.C. Bonamy %A T. Wu %A S.W. Barnes %A S.E. Bopp %A R. Borboa %A A.T. Bright %A J. Che %A S. Cohen %A N.V. Dharia %A K. Gagaring %A M. Gettayacamin %A P. Gordon %A T. Groessl %A N. Kato %A M.C.S. Lee %A C.W. McNamara %A D.A. Fidock %A A. Nagle %A T-g Nam %A W. Richmond %A J. Roland %A M. Rottmann %A B. Zhou %A P. Froissard %A R.J. Glynne %A D. Mazier %A J. Sattabongkot %A P.G. Schultz %A T. Tuntland %A J.R. Walker %A Y. Zhou %A A. Chatterjee %A T.T. Diagana %A E.A. Winzeler %J Science %D 2011 %O DOI:10.1126/science.1211936
-
Interest in Links to Patents From Structures in ChEMBL
We are exploring establishing links from the ChEMBL compounds to patents. The implementation can have two basic routes....
- Links from the interface to patents (simple and quick to do now we have UniChem).
- Patent uri's in the database itself (more complex, and more difficult to keep up to date, but arguably more useful).
So to help our planning for next year, comments, wishes are most welcome.... -
New Drug Approvals 2011 - Pt. XXXI Asparaginase Erwinia chrysanthemi (ErwinazeTM)

ATC code: L01XX02
On November 18, the FDA approved asparaginase from Erwinia chrysanthemi for the treatment of patient with acute lymphoblastic leukemia (ALL) who have become allergic to the E. coli asparaginase that is conventionally used for the treatment of ALL patients.
ALL is a cancer of the white blood cells and can be fatal within weeks from the onset of the disease if it is left untreated. In ALL, there is an unproportional increase in the population of immature white blood cells, which crowd out functional immune cells as well as red blood cells and platelets, and in advanced stages of the disease infiltrate into tissues and organs, most frequently liver, spleen and lymph nodes. The symptoms of ALL in its initial stages are fatigue, anemia, frequent infections and fever as well as breathlessness and prolonged bleeding. ALL is caused by DNA damage and associated with exposure to radiation and cancerogenic chemicals. There are a number of typical chromosomal translocations, the most frequent in adults with ALL is the so called Philadelphia chromosome, where a translocation of chromosomes 9 and 22 results in the formation of the fusion gene bcr-abl (CHEMBL1862). Treatment of ALL is usually encompasses the three phases i) remission induction, which aims to quickly kill off 95% of the cancerous cells, ii) consolidation to further reduce the tumor burden and iii) maintenance with the aim to prevent a relapse caused by stray surviving leukemic cells.
Aspariginase is part of the remission induction regimen and used in combination with other cytostatic and cytotoxic drugs including prednisolone (CHEMBL131), dexamethasone (CHEMBL384467) , vincristin (CHEMBL303560) and daunorubicin (CHEMBL178). Asparaginase is an enzyme (EC 3.5.1.1) that catalyzes the hydrolysis of asparagine (CHEBI:17196) to aspartic acid (CHEBI17053) as shown below. Unlike healthy body cells, leukemic cells rely on the presence of extracellular asparagine for protein metabolism and survival, hence the beneficial effect for ALL patients. Conventionally, asparaginase for he treatment of ALL is derived from the bacterium Escherichia coli (corresponds to Uniprot:P00805) but some patients (estimated 10-15%) become allergic to this enzyme. For these patients, the ortholog from the bacterium Erwinia crysanthemi (Uniprot-Id P06608) is available to replace the untolerated treatment.

Asparaginase catalyzes the hydrolysis of asparagine to aspartic acid. Asparaginase Erwinia crysanthemi is a tetrameric enzyme composed of four identical subunits each weighing about 35kDa. The crystal structures of many asparaginase enzymes are known, including that of the closely related protein from Erwinia carotovora (PDBe:2jk0)
The recommended dosage is 25.000 International Units/m2 (International Units compare biological activity relative to an arbitrary amount of the active ingredient, square meter refers to body surface) three times a week. The route of administration is intramuscular injection.
The pharmakokinetic paramaters of Erwinaze were not determined in clinical trials, serum concentrations greater than 0.1 International Units/mL were reached by all patients within 72 hours of the third injection of aspariginase Erwinia chrysanthemi.
Side effects reported in the study include serious hypersensitivity reactions, pancreatitis, glucose intolerance, thrombosis and hemorrhage.
Asparaginase Erwinia chrysanthemi is marketed in the US by Eusa Pharma under the trade name Erwinaze. In a number of other countries, it is available under the name Erwinase.
Package information can be found here.
-
New Drug Approvals 2011 - Pt. XXX - Aflibercept (EyleaTM)
 On November 18th 2011, the FDA approved Aflibercept (trade name: Eylea; Research Code: AVE-0005, also known as VEGF Trap), a recombinant fusion protein indicated for the treatment of patients with neovascular (wet) age-related macular degeneration (AMD).
On November 18th 2011, the FDA approved Aflibercept (trade name: Eylea; Research Code: AVE-0005, also known as VEGF Trap), a recombinant fusion protein indicated for the treatment of patients with neovascular (wet) age-related macular degeneration (AMD).
AMD is an eye condition which usually occurs in older patients and affects the macula area of the retina, causing loss of vision and eventually blindness. In particular, wet AMD is characterised by an abnormal growth of new blood vessels (neovascularisation) behind the retina. This originates from an abnormal activation of angiogenesis, by the vascular endothelial growth factor-A (VEGF-A; ChEMBL: CHEMBL1783; Uniprot: P15692) and the placenta growth factor (PlGF; ChEMBL: CHEMBL1697671; Uniprot: P49763), of the vascular endothelial growth factor receptors VEGFR-1 (ChEMBL: CHEMBL1868; Uniprot: P17948) and VEGFR-2 (ChEMBL: CHEMBL279; Uniprot: P35968), two receptor tyrosine kinases present on the surface of endothelial cells. This leads to abnormal increased permeability, scarring and possibly to the loss of fine-resolution central vision. Aflibercept acts as a soluble 'decoy' receptor that binds VEGF-A and PlGF and thereby inhibits the binding and activation of the VEGFR-1 and VEGFR-2 receptors.
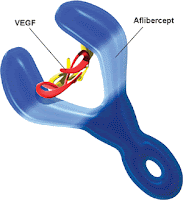 Aflibercept is a recombinant fusion protein that incorporates portions of extracellular domains of the human VEGFR-1 (containing Ig-like C2-type 2 domain fragment; Uniprot: P17948|151-214|) and VEGFR-2 (containing Ig-like C2-type 3 domain fragment; Uniprot: P35968|224-320|) fused to the Fc portion of human immunoglobulin G1 (IgG1). Aflibercept is a dimeric glycoprotein with a protein molecular weight of 97 kDa (115 kDa with glycosylation).
Aflibercept is a recombinant fusion protein that incorporates portions of extracellular domains of the human VEGFR-1 (containing Ig-like C2-type 2 domain fragment; Uniprot: P17948|151-214|) and VEGFR-2 (containing Ig-like C2-type 3 domain fragment; Uniprot: P35968|224-320|) fused to the Fc portion of human immunoglobulin G1 (IgG1). Aflibercept is a dimeric glycoprotein with a protein molecular weight of 97 kDa (115 kDa with glycosylation).
>Aflibercept SDTGRPFVEM YSEIPEIIHM TEGRELVIPC RVTSPNITVT LKKFPLDTLI PDGKRIIWDS RKGFIISNAT YKEIGLLTCE ATVNGHLYKT NYLTHRQTNT IIDVVLSPSH GIELSVGEKL VLNCTARTEL NVGIDFNWEY PSSKHQHKKL VNRDLKTQSG SEMKKFLSTL TIDGVTRSDQ GLYTCAASSG LMTKKNSTFV RVHEKDKTHT CPPCPAPELL GGPSVFLFPP KPKDTLMISR TPEVTCVVVD VSHEDPEVKF NWYVDGVEVH NAKTKPREEQ YNSTYRVVSV LTVLHQDWLN GKEYKCKVSN KALPAPIEKT ISKAKGQPRE PQVYTLPPSR DELTKNQVSL TCLVKGFYPS DIAVEWESNG QPENNYKTTP PVLDSDGSFF LYSKLTVDKS RWQQGNVFSC SVMHEALHNH YTQKSLSLSP G
Other therapies to treat AMD are available on the market and these include Verteporfin (ChEMBL: CHEMBL1200573; approved in 2000; trade name Visudyne), Pegaptanib sodium (ChEMBL: CHEMBL1201421; approved in 2004; trade name Macugen) and Ranibizumab (ChEMBL: CHEMBL1201825; approved on 2006; trade name Lucentis).
Aflibercept recommended dosage is 2 mg administrated by intravitreal (into the eye cavity) injection every 4 weeks for the first 12 weeks, followed by 2 mg via intravitreal injection once every 8 weeks.
Following intravitreal administration of 2 mg per eye, a fraction of the administrated dose binds to the endogenous VEGF in the eye to form the inactive Aflibercept:VEGF complex. Once absorbed into the systemic circulation, Aflibercept presents in the plasma as the free unbound Aflibercept and predominantly as the inactive Aflibercept:VEGF complex. Aflibercept has a volume of distribution (Vd) of 6 L and a terminal elimination half-life (t1/2) of 5 to 6 days after iv administration of doses of 2 to 4 mg.kg-1 of Aflibercept. Aflibercept undergoes elimination through both target-mediated disposition via binding to free endogenous VEGF and metabolism via proteolysis.
The full prescribing information for Eylea can be found here.
The license holder is Regeneron Pharmaceuticals, Inc. and the product website is www.eylea.com. -
New Drug Approvals 2011 - Pt. XXIX (ruxolitinib phosphate) (Jakafi ™)
ATC Code: L01XE18
Wikipedia:Ruxolitinib
Myelofibrosis is a disorder of the bone marrow, in which the marrow is replaced by scar (fibrous) tissue. Scarring of the bone marrow reduces its ability to blood cells, and can lead to anemia, bleeding problems, and a higher risk of infections due to reduced white blood cells. It is also associated with engorgement of organs suchs as the spleen and liver. Primary myelofibrosis may develop to secondary myelofibrosis - including leukemia and lymphoma. Myelofibrosis is associated with dysregulated Janus kinases JAK1 and JAK2, and some with a somatic mutation in JAK2 (JAK2V617F) (OMIM). JAK signaling involves recruitment of STATs (signal transducers and activators of transcription) to cytokine receptors, activation and subsequent localization of STATs to the nucleus leading to modulation of gene expression. Oral administration of ruxolitinib prevented splenomegaly, preferentially decreased JAK2V617F mutant cells in the spleen and decreased circulating inflammatory cytokines.
JAK1 (Uniprot:P23458) and JAK2 (Uniprot:O60674) are tyrosine protein kinases and members of the Janus kinase subfamily, where all members of the family contain two tandem protein kinase domains (PFAM:PF00069), one of which is catalytically active and one believed to be inactive. JAK1 and JAK2 are 43% identical by sequence and both have the 3D structure of their kinase domain determined (see e.g. PDBe:3EYH and PDBe:3Q32 for JAK1 and JAK2 respectively). Ruxolitinib is the first approved targeted JAK inhibitor, with several others in mid to late-stage clinical development (including CYT-387, GLPG-0634, INCB-28050, ONX-0803, NS-018, pacritinib (SB-1518), AZD-1480, BMS-911543, LS-104, XL-019, TG-101348, tofacitinib (CP-690550), VX-509, R-348, WHI-P131 and oclacitinib (PF-03394197) (veterinary applications)) - note these show a broad range of selectivity against the three known JAK subtypes.
Ruxolitinib (IUPAC: (R)-3-(4-(7H-pyrrolo[2,3 d]pyrimidin-4-yl)-1H-pyrazol-1-yl)-3-cyclopentylpropanenitrile phosphate; (Standard InCHI key: HFNKQEVNSGCOJV-OAHLLOKOSA-N) has a molecular weight of 404.36, an AlogP of 2.88 and complies with all components of Lipinski's rule of 5.
Ruxolitinib is administered orally as the phosphate salt as tablets and dosed according to platelet count (hence the large range of dosage forms). Each tablet contains ruxolitinib phosphate equivalent to 5 mg, 10 mg, 15 mg, 20 mg and 25 mg of ruxolitinib free base. The Tmax for Ruxolitinib is 1-2 hours post dosing, with exposure (Cmax and AUC) linear over a dose range of 5mg to 200mg. Oral absorption is in excess of 95%. The volume of distribution is 53-65 L, with plasma protein binding in excess of 97%. Ruxolitinib is predominantly metabolized by CYP3A4, with the two primary metabolites displaying weaker, but still significant pharmacological activity against their specific targets. Administration of ruxolitinib with ketoconazole, a potent CYP3A4 inhibitor, prolongs the half life of ruxolitinib from 3.7 to 6.0 hours, increases the Cmax to 33% and the AUC to 91%. The change in the pharmacodynamic marker, pSTAT3 inhibition, was consistent with the corresponding ruxolitinib AUC following concurrent administration with ketoconazole.
The license holder for Jakafi is Incyte, and the full prescribing information can be found here.








