-
ChEMBL Goes to A Conference - Fall ACS 2009
 We are going to speak at the 238th ACS National Meeting, Washington, DC, August 16-20, 2009, on "ChEMBL: Large-scale Mapping of Medicinal Chemistry and Pharmacology Data to Genomes". The Abstract for the talk is:
We are going to speak at the 238th ACS National Meeting, Washington, DC, August 16-20, 2009, on "ChEMBL: Large-scale Mapping of Medicinal Chemistry and Pharmacology Data to Genomes". The Abstract for the talk is:
Although the majority of effective therapeutics are small molecules, there is relatively little readily accessible public domain data mapping drugs to their molecular targets. When one considers clinical trial stage, or discovery stage data, the situation deteriorates further. However, this type of data is essential for Chemical Biology experiments, and is crucial for informed target selection in drug discovery. To address this issue, we have built a series of large scale databases, known as ChEMBL, that map small molecule structures to their target genes and also their functional effects. This data also captures a large ammount of human and model organism pharmacological data, systems often used in pre-clinical validation and safety pharmacology testing. A variety of applications of these databases in the area of target prioritisation, lead discovery, lead optimisation and drug repurposing will be described.
-
Books and Papers - 8 - Programming Collective Intelligence
 I am on holiday today - sort of. Went to Borders for a Starbucks (product placement hopefully pays well), and while queuing for my Orange Mocha Frappuccino!, I caught sight of the O'Reilly books; one stood out from the crowd - Programming Collective Intelligence. It looks a very cool collection of code (Python) implementing a whole variety of data analysis/machine learning techniques and routines to build smarter, more responsive and adaptive web 2.0 applications. Skimming the pages while I had my caffeine speedball led me to spend my cash.
I am on holiday today - sort of. Went to Borders for a Starbucks (product placement hopefully pays well), and while queuing for my Orange Mocha Frappuccino!, I caught sight of the O'Reilly books; one stood out from the crowd - Programming Collective Intelligence. It looks a very cool collection of code (Python) implementing a whole variety of data analysis/machine learning techniques and routines to build smarter, more responsive and adaptive web 2.0 applications. Skimming the pages while I had my caffeine speedball led me to spend my cash.
%A Toby Segaran %T Programming Collective Intelligence %I O'Reilly %D 2007 %O ISBN 978-0596529321
-
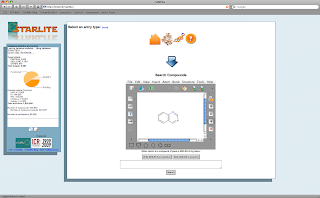
One StARlite interface
No, this isn't a hotel review, despite the picture above; however since finding this image, I now feel compelled to visit Carlsbad, NM to pose under the sign. Now for the post itself; find below some screenshots for an internal interface for StARlite data developed by our close collaborators at the Institute of Cancer Research in Sutton. This interface for StARlite shows some basic workflow themes that give some ideas as to the potential uses of StARlite 'straight out of the tin'. Several of the views will be incorporated into the EMBL-EBI public web interface ;) Bissan's group is developing an integrated system for cancer chemogenomics, called canSAR.
Compound Searching: What would an SAR database be without a compound sketcher and search mechanism. Well here is one, implemented with Marvin, and the Dotmatics Pinpoint cartridge.
Compound Browsing: A "Top Trumps" view on compounds is quite a useful paradigm for browsing and selecting compounds for further analysis. As an aside, for an example of sensory overload on a web page, try out the current Top Trumps company website.
 Target Browsing: It is useful sometimes to browse the target dictionary to explore its structure, coverage, and to pre-group compounds into various activity sets.
Target Browsing: It is useful sometimes to browse the target dictionary to explore its structure, coverage, and to pre-group compounds into various activity sets.
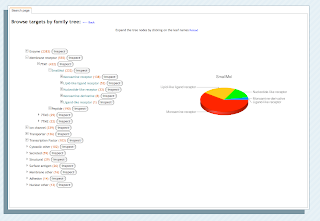 This page uses the Google Chart API to do some of the heavy lifting. In which the chart itself is made using a URL of the form...
This page uses the Google Chart API to do some of the heavy lifting. In which the chart itself is made using a URL of the form...
http://chart.apis.google.com/chart?cht=p3&chd=t:60,40&chs=250x100&chl=Good|Evil
Which produces the following (and you thought Google could do no Evil).
I thank Mark Halling-Brown, and Bissan Al-Lazikani of the ICR for the images. However, I do not thank them for the geek joke in the use of the StarWarsTM font for StARlite (Ha, ha, ha, ha, ha.....).
-
Books and Papers - 7 - Population Genetics, Molecular Evolution, and The Neutral Theory by Motoo Kimura
 Published in 1994 in recognition of the huge influence of Motoo Kimura on the field of theoretical studies on molecular evolution. This is a collection of papers and essays published by Kimura over the period 1955 to 1986. The writing is just truly beautiful, the prose, pace and clarity in the text humbles me as a supposed native English speaker (as this blog so clearly shows!). If you're not interested in the science at all, just buy it for the masterclass of technical writing inside.
Published in 1994 in recognition of the huge influence of Motoo Kimura on the field of theoretical studies on molecular evolution. This is a collection of papers and essays published by Kimura over the period 1955 to 1986. The writing is just truly beautiful, the prose, pace and clarity in the text humbles me as a supposed native English speaker (as this blog so clearly shows!). If you're not interested in the science at all, just buy it for the masterclass of technical writing inside.
The theme of the book is around The Neutral Theory, quite a contentious issue in evolution (essentially, this states that the vast majority of observed mutations at a molecular level are not adaptive; now flame me!) This book changed the way I thought about mutation, protein sequence and structure and function. Forever.
%T Population Genetics, Molecular Evolution, and The Neutral Theory %A Motoo Kimura %E J.F. Crow %D 1994 %I Chicago %O ISBN: 0-226-43562-8
-
Protein Ligand Affinity Databases
 I came across a nice tabular summary of some existing 'public' 'primary' protein-ligand interaction databases that primarily focus on protein-ligand affinity data (so Ki, Kd, IC50, EC50, etc.), that I have reproduced below (many thanks to Helena Strömbergsson, from Uppsala University for the data).
I came across a nice tabular summary of some existing 'public' 'primary' protein-ligand interaction databases that primarily focus on protein-ligand affinity data (so Ki, Kd, IC50, EC50, etc.), that I have reproduced below (many thanks to Helena Strömbergsson, from Uppsala University for the data).
Name Target Class Focus Approx size BindingDB All ~48,000 PDSP Receptors ~47,000 BRENDA Enzymes ~19,000 BindingMOAD All ~3,500 PDBBind All ~3,500 AffinDB All ~700 PLD All ~500 The comparable number from StARlite (31) are 507,645 (of which 186,370 are better than 100nM) for affinity class end-points. Oh, and we have started a new load.....
-
Parallel Sets for Drug Delivery Class and RoF
 So, following on from the post of a few days ago is the following diagram. It shows, in parallel sets form, the proportion of drug structures (the dosed components) that either fail or pass the rule of five (RoF) - in this case a molecule fails if it fails any single rule, compared to the delivery route of the drug, in this case Oral, Parenteral and Topical. As always, things are not as simple as they seem, since some drugs are dosed both topically, and orally and parenterally, and so forth. By comparison to the example for the Titanic, this would be the same as a passenger being both First Class and Second Class at the same time. For clarity, here are some summary numbers 68% of RoF compliant drugs can be Orally dosed, compared to 50% of RoF 'uncompliant' drugs that can be orally dosed. But I think the graph is interesting nonetheless.
So, following on from the post of a few days ago is the following diagram. It shows, in parallel sets form, the proportion of drug structures (the dosed components) that either fail or pass the rule of five (RoF) - in this case a molecule fails if it fails any single rule, compared to the delivery route of the drug, in this case Oral, Parenteral and Topical. As always, things are not as simple as they seem, since some drugs are dosed both topically, and orally and parenterally, and so forth. By comparison to the example for the Titanic, this would be the same as a passenger being both First Class and Second Class at the same time. For clarity, here are some summary numbers 68% of RoF compliant drugs can be Orally dosed, compared to 50% of RoF 'uncompliant' drugs that can be orally dosed. But I think the graph is interesting nonetheless.
As you know by now, I am completely incapable of doing any actual real work or data analysis, so the keyboard wizard in this case was Bissan Al-Lazikani.
So I must admit, I don't find Parallel Sets a compelling way to view these data (maybe the colors, maybe because the trends aren't that clear?).
-
Set Visualisation
 A quick link to a page on the BBC website. It (parallel sets) is a nice visual way of presenting the association of various categories. Here is a link to another example of parallel sets culled from the interweb.
A quick link to a page on the BBC website. It (parallel sets) is a nice visual way of presenting the association of various categories. Here is a link to another example of parallel sets culled from the interweb.
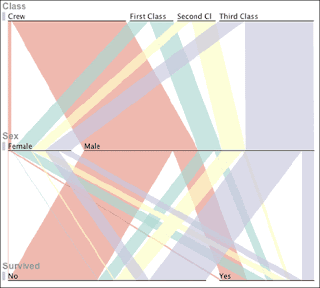
The example used on the BBC web site (and illustrated below) is for the relationship of gender, ticket class (or crew), and survival following the sinking of the Titanic. I will try and put together something for drugs (oral, topical, parenteral) and rule-of-five classification and post it here, see what it looks like. Maybe I will try and partition our set of development candidates into launched and failed as well to make it more interesting, also prodrugs,......
-
Privileged Scaffold Identification from StARlite data
 Here is a little use-case of StARlite, I doubt we will ever formally publish it, but hopefully it is quite interesting nonetheless.
Here is a little use-case of StARlite, I doubt we will ever formally publish it, but hopefully it is quite interesting nonetheless.
A common requirement in hit discovery is the selection of a set of compounds likely to be bioactive against a particular target or target family (for example a protein kinase focussed library, etc), there are many ways of doing this task, and for screening a new target this sort of approach is often very economically cost-effective. Usually there is the concept of a set of active series, or chemotypes for a target family, and this is often expressed as a 'scaffold' - the chemical core - often a rigid part of a molecule with all the surface 'decoration' removed. More particularly, this 'scaffold' is often associated with some synthetic accessibility concepts for use in library synthesis, parallel chemistry and so forth. Anyway, forgetting some of the details and complexities of deciding 'what is a series', 'what is a scaffold', etc. here is an annotated workflow.
- Identify all scaffolds within StARlite (we use Pipeline pilot for this, but there are many programs that can do this).
- Build a table of the presence of each scaffold within each molecule (remember some scaffolds will be substructures of others, a molecule can contain several distinct scaffolds, etc.
- Identify an Active Set within StARlite (it can be those active against a gene family, say rhodopsin-like GPCRs, a particular target, CNS penetrant compounds, compounds with long half-lives, it doesn't really matter). Of course, you also need to assign an activity cutoff (we tend to use 1 micromolar).
- Assign all molecules in StARlite to either the Active Set (those in 3) or an Inactive Set (all other molecules).
- Do some simple set manipulations on the scaffolds preferentially active within the active set. (look for rank or proportion enrichment, etc).
- Rank the enriched scaffolds by some criteria (we like to use a ligand efficiency proxy (dividing the enrichment ratio by the molecular weight/number of heavy atoms).
In practice, the active and pseudo-inactive sets can be used to build far more sophistacted activity classifiers, pharmacophores, etc.
Let's look at a toy example.
We did a blast query on StARlite using the sequence of human MCH-1 as a query. This pulls out a list of targets ranked by similarity to MCH-1, we selected a set of similar sequences as our MCH-1-like active set, and extracted all molecules and assays assigned to this set of sequences. We then looked at the scaffold frequency rank within the active set (dark blue) compared to the rank of the scaffold of the entire database (light blue). We then computed the enrichment ratio of the active set to the entire database (Fragment Specificity, in red). A non-enriched scaffold will show a value close to zero, the higher the degree of association, the higher the score. Finally, we 'normalised' the relative enrichment by molecular weight of the scaffold (Fragment Elegance, light green).
So overall, we identify a set of scaffolds that are biased towards a particular activity class - in this case, scaffolds 6 and 7 (dark blue) are probably the best, whilst 8, although more associated with MCH-1-like activity (a higher red score) are less attractive due to the inherently higher molecular weight (and synthetic access difficulties) of this particular scaffold.