-
A Dating Site For Chemists and Biologists
Probably everyone who reads the ChEMBL-og will have world-changing ideas - but it's really difficult to find someone to screen a few compounds for you - of course there are CROs who will want to meet, then prepare a quote for you, set up a CDA, receive payment, etc., but cash is difficult to get hold of, and the process will be slow. There are no grant mechanisms for this sort of thing either - imagine - "I'd like funds to test four compounds as potential inhibitors of snoraze" - no chance (at least with the panels I've sat on) too small, too speculative.... The bigger problem though is finding someone with the assay or the compounds.But, there's a lot of people with compounds to test, and a lot of biologists with assays that are easy to run in their labs, and they have expertise in, but who can't assemble sets of interesting compounds to profile. Why not just use the paradigm of a dating site to matchmake mutually compatible biologists and chemists - if there is a spark, it could develop into a long lasting (collaborative) relationship!Imagine something like:Biologist with HMGCoA reductase assay and expertise in cholesterol homeostasis would like to meet chemist with non-statin compounds likely to be brain penetrant to test a cool idea.
Anyway, there's a toy FaceBook group that I've set up - just to get the idea across. I've pitched this as a national thing (so for me that means to the UK, for you somewhere different maybe) - not least that it's a lot easier to ship compounds around within a country than between - and also there's a clear match to downstream funding opportunities. I chose FaceBook, since most of the open LinkedIn groups I'm involved in are train-wrecks of spam and flame-wars.I think this idea is worth trying, or at least getting some discussion started over - huge thanks to Tom Heightman for our recent discussion on things that needed to be done in Chemical Biology in the UK.Maybe Google+ is another alternative.
-
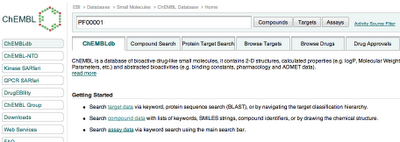
Pfam domain searching of targets in ChEMBL
One thing new in the backend and interface for this release of ChEMBL is the ability to search for targets containing particular PFAM domains. So if you know a PFAM id, you can search in the search box (and then select "Targets" for that domain. For example, PF00001 is the Pfam ID for the rhodopsin-like GPCRs.
A couple of important things on this though - the current functionality does exactly what it says - it returns proteins that contain that domain - the compounds do not necessarily (and often in fact do not) bind at that domain. This multidomain, and multi protein target issue is a surprisingly big challenge, and is a big trap for the unwary. So caveat emptor.
We do plan in the next release or two, provide a prediction of the likely/known compound binding domain (however here, for proteins that contain multiple copies of the predicted/lknown binding domain it is complicated....). -
ChEMBL 13 Released
We are pleased to announce the release of ChEMBL_13. This latest version of the ChEMBL database contains:- 1,296,266 compound records
- 1,143,682 distinct compounds
- 617,681 assays
- 6,933,068 bioactivities
- 8,845 targets
- 44,682 documents
- 8 data sources
Please refer to the ChEMBL_13 release notes for a more detailed description of all changes included in this release.
We have also made a couple of minor updates to the interface which include- New Ligand Efficiency widget, which is displayed on the Target report card pages (e.g. CHEMBL331)
- Added external links to Pfam, Array Express and Human Protein Atlas on the Target report card pages
- Added external links to ATC/DDD Index on Compound report card pages (e.g. CHEMBL1642)
You can download the data from the ChEMBL ftpsite: ftp://ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/latest/ -
PPI-Net - UK National Network for Collaboration on Protein-Protein Interactions
Yesterday I took part in a workshop on targeted libraries at the Univeristy of Leeds - it was a great meeting, with lots of good ideas on directing compound design towards this class of interaction (Protein-Protein Interactions, PPIs - now form a significant fraction of the interesting target systems for drug discovery and basic biology and the development of tool compounds/leads that modulate these is a major challenge due to the low ligand efficiency characteristics of the majority of PPI binding sites).
There is a great website, with a collection of key references, contacts, etc.
I came away determined to do a couple of things:
- Resurrect some old F77 code (yay!) to look at peptide binding pharmacophores.
- Try and play a bit with Facebook/LinkedIn as a sort of dating site for unattached chemical/biologists.
-
ACS meeting in San Diego
Three of the group (Patricia Bento, Jon Chambers and I) are out at the ACS National in San Diego at the end of March. If you want to meet up and have a coffee, get in touch, if you'd like a talk on ChEMBL, I'm sure we could fit something in. -
Paper: Toxicogenomics Investigation Under the eTOX Project
A paper on some of the toxicogenomics work we are involved in under the IMI eTox project. The paper is Open Access :) and downloadable here.
%T Toxicogenomics Investigation Under the eTOX Project %A O. Taboureau %A A. Hersey %A K. Audouze %A L. Gautier %A U.P. Jacobsen %A R. Akhtar %A F. Atkinson %A J.P. Overington %A S. Brunak %O http://dx.doi.org/10.4172/2153-0645.S7-001 %J Pharmacogenomics & Pharmacoproteomics %V S7 %D 2012
-
Sequence-Structure alignment of the 11 structurally characterised distinct GPCRs
Here is a joy formatted alignment of the (now) 11 sequence distinct rhodopsin-like GPCR structures - I've selected a representative for those for which there are multiple structures known - usually those that are most complete in terms of lack of disordered loops, etc. The alignment is quite unstable in parts, and several regions are open to interpretation.....
The structures are:
- 3uon - human muscarinic M2 receptor
- 4daj - rat muscarinic M3 receptor
- 3rze - human histamine H1 receptor
- 2rh1 - human beta-2 adrenergic receptor
- 2vt4 - turkey beta-1 adrenergic receptor
- 3pbl - human dopamine D3 receptor
- 2ydv - human adenosine A2a receptor
- 3v2w - human sphingosine-1-phosphate receptor
- 3odu - human CCR4 receptor
- 2i35 - bovine rhodopsin
- 2z73 - squid rhodopsin
The next to be released structures will almost certainly be the mu-opioid receptor (PDB code 4DKL) and the kappa-opioid receptor (PDB code 4DJH), which are on hold awaiting publication.
10 20 30 40 50 3uon ( 20 ) tfevvfivl 4dajA ( 64 ) iwqvvfiaf 3rze ( 28 ) mplvv 2rh1 ( 29 ) devwvvgmgi 2vt4A ( 40 ) weagmsl 3pblA ( 32 ) yal 2ydv ( 3 ) imgssvYit 3v2w ( 17 ) sdyvnydIIvrHYnyTgklnisa ltsv 3oduA ( 27 ) pçfre-------------------------enanfnkiflpt 1u19A ( 1 ) mnGtegpnfyVPfsnktgvVrsPFeapQyyLaepwqFsmlAa 2z73A ( 9 ) etwwyNpsIvVhpHWref--------------dqvpdavYyslGi aaaaa 60 70 80 90 100 3uon ( 29 ) vagslSlvTiigNilVmvSIkvnrhLqtvnnyflfSLAcADliiGvfSMn 4dajA ( 73 ) ltgflAlvTiigNilVivAFkvnkqLktvnnyFllSLAcADliIGviSMn 3rze ( 33 ) vlsticlvTvglNllVlyAvrserkLhtvGnlYIvsLSvADliVGavVMp 2rh1 ( 39 ) vmslivlaIvfgNvlVitAIakferLqtvtnyFItsLAcADlvMGlaVVp 2vt4A ( 47 ) lmalVvllIvagNvlViaAigstqrLqtltnlFItsLAcADlvvGllVVp 3pblA ( 35 ) sYcalilaIvfgNglVcmAVlkeraLqtttnyLVvsLAvADllvAtlVMp 2ydv ( 12 ) vElaiavlAilgNvlVcwAvwlnsnLqnvtnyFVvsAAaADilVGvlAIp 3v2w ( 51 ) vfiliCcfIileNifvlltiwktkkFhrpMYyFIgnLAlSDllaGvaYta 3oduA ( 44 ) iYsiIfltGivgNglvilvMgyqkklrsmtdkYRlhLSvADllFVitLpf 1u19A ( 43 ) yMflLimlGfpiNflTlyVTvqHkkLrtplNyILlnLAvADlfMVfg-GF 2z73A ( 40 ) fIgiCgiiGcggNgiViyLFtktksLqtpanmFiinLAfSDftFSlvNGf aaaaaaaaaaaaaaaaaaaaaa aaaaaaaaaaaaaaaaaa aaa 110 120 130 140 150 3uon ( 79 ) lytlytvi-gyWplgpvvÇdlWlalDYvVSNAsVmNLliiSfdryfcvtk 4dajA ( 123 ) lFttyiim-nrWalgnlaÇdlwLSiDYvASNAsVmNLlvISfDryfsitr 3rze ( 83 ) mnilyllm-skwsLgrplÇlfWLSmDYVASTASIfSVfiLCiDryrsvqq 2rh1 ( 89 ) fgaahilm-kmWtfgnfwçefWTSiDVlCVTASIeTLcvIAvdryfAIts 2vt4A ( 97 ) fgatlvvr-gtWlwgsflçelWTSlDVlCVTAsIeTLcvIAiDrylaits 3pblA ( 85 ) wvvylevtggvWnfsricÇdvFVTlDVmMcTAsIwNLCaISidRytAVvm 2ydv ( 62 ) faiaIst---GfçaaçhgÇLfiACfVLVLTASSIfSLlaIAiDryiairi 3v2w ( 101 ) Nlllsga--tTykLtPaqWFlREGsMFvALSASVfSLlaIAieryitmlk 3oduA ( 94 ) WavDAva---nWyfgnflÇkaVHviYTVNlYSSVwILAfISlDRylAiVh 1u19A ( 92 ) tTTlyTSlhGyFvfgptGÇnlEGffATLGGEIaLWSLvvLaieRyvvVck 2z73A ( 90 ) plMtiSCflkkWifgfaaÇkvYGfiGGiFGFMsIMTMAMiSiDrynViGr aaaaaaa aaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa 160 170 180 190 200 3uon ( 128 ) pltypvk---rttkmAgmmiaaAwvlSfilwapaIlfwqfivg------- 4dajA ( 172 ) pltyrak---rttkrAgvmiglAwviSfvlWApaIlfwqyfvg------- 3rze ( 132 ) plrylky---rtktrAsatilgawflSfl-WvipIlgwnh 2rh1 ( 138 ) pfkyqSl---ltknkArviilmvwivSgltSflpIqmhwyr-----athq 2vt4A ( 146 ) pfryqsl---mtrarAkviictvwaiSalvSflpImmhwWr-----dedp 3pblA ( 135 ) pvhyqhgtgqsscrrValmitavwvlAfaVSc-pLlfgfNtTg------- 2ydv ( 109 ) plryngl---vtgtrAkgiiaicwvlSfaIGltPmlgwnnÇgqp--kegk 3v2w ( 156 ) nnfrlfllisacwviSlilGglPimgwn----------- 3oduA ( 141 ) atn----sqrprkllAekvVyvgVwipAlllT-ipDfif-Anvsead--- 1u19A ( 142 ) pmsn----frfgenhaimgvafTwvmAlaCAapPlvgwSrYIPE------ 2z73A ( 140 ) pmaas---kkMshrrAfimiifVwlwSvlwAigPifgwGaYtLE------ aaaaaaaaaaaaaaaaaaa 210 220 230 240 250 3uon ( 168 ) ----vrtVedgeÇyIqff------snaavtfgtAiaaFylpviiMtvlyw 4dajA ( 212 ) ----krtVppgeÇfIqfl------septitfgtAiaaFymPvtiMtilyw 3rze ( 175 ) rredkÇeTdfy------dvtwfkvmtaiinFylPtllMlwfya 2rh1 ( 180 ) eAinÇyae-etçÇdff--------TnqayaiasSivSFyvplviMvfvYs 2vt4A ( 188 ) qAlkçyqd-pgçÇdfv--------TnrayaiasSiiSFyipLliMifval 3pblA ( 177 ) --------dptvÇsIs---------npdFViySSvvSFylPfgvTvlvya 2ydv ( 154 ) ahsqgÇgegqvAÇlFedVV-----pmnYMVyfNffaCVlvPlllMlgvyl 3v2w ( 184 ) ----ÇisalssÇSTVLP-------LYhkhYIlfCTtvFtllllsIvilYc 3oduA ( 182 ) --------dryiÇdrfyp---ndlwvvvfqfqhimvglilPgivIlsCyc 1u19A ( 182 ) -------GMQCSÇGIDYYTpheetnNesFViyMfvvHfiiPlivIffcyg 2z73A ( 181 ) -------GVLCNÇSFdYIsr--dsttrsNIlcMFilGffgPiliiffCyf aaaaaaaaaaa aaaaaaaaaaaa 260 270 280 290 300 3uon ( 208 ) hisrasksri pppsrekkvtrtilaIllaFi 4dajA ( 252 ) rIyketek like aqTlsaIllaFi 3rze ( 212 ) kIykaVrqhc lhmnrerkaakQLgfIMaaFi 2rh1 ( 221 ) rVfqeakrql kfclkeHkaLktlgiIMgtFt 2vt4A ( 229 ) rvyreakeq irehkalktlgiImgvFt 3pblA ( 210 ) rIyvvlkqrrrk-----------------gvplrekkatqMVaiVlgaFi 2ydv ( 199 ) rIflaarrqlkqmesq stlqkevhaakSLaiIvglFa 3v2w ( 223 ) riyslvrtr asrssenvaLlkTViiVLsvFi 3oduA ( 221 ) iIisklshs kghqkrkalktTviLilaFf 1u19A ( 225 ) qLvftvkeaaaq------------qqesattqkaekevTrMviiMviaFl 2z73A ( 222 ) nIvmsvsnhekemaamakrlnakelrkaqaganaemrlAkIsivIVsqFl aaaaa aaaaaaaaaaaaaaaaa 310 320 330 340 350 3uon ( 398 ) itWapYNvmVlintfçap--------ç--ipntvwtiGywlCYinstiNp 4dajA ( 501 ) itWtpyNimVlvntfçds--------ç--ipktywnlgywlCYiNStvNP 3rze ( 426 ) lCWipYFiffmviafçkn--------ç--cnehlhmftiWlGYiNStlNP 2rh1 ( 284 ) lcWlpFFiVNivhviqdn----------lirkevyillNwiGYvNSgfNp 2vt4A ( 301 ) lCWlpFFlvnivnvfnrd----------lvpdwlfvafnwlGYAnSAmnp 3pblA ( 340 ) vCWlpFFltHvlnthçqt--------ç-hvspelysattwlGYvNsalNP 2ydv ( 244 ) lCWlpLHiiNcftffçpd--------çshaplwlMylAivlSHtNSvvNP 3v2w ( 267 ) acwapLFiLLllDvgçkvk------tç--diLfrAeyfLvlAvlNSgtNP 3oduA ( 250 ) acWlpyyigisidsfilleiikqgçefentvhkwisitEAlAFfHCclNp 1u19A ( 263 ) iCWlpYAgvAfyIfthqgsd---------fgpifMTipAFfAKtSAvyNP 2z73A ( 272 ) lSWspYAvvAllAQfgplew---------VtpyaAQlpVMfAKaSaihNP aaaaaaaaaaaaaaa aaaaaaaaaaaaa aaa 360 370 380 390 400 3uon ( 438 ) acYalcnatFkktfkhllm 4dajA ( 541 ) vcYalcnktFrttfkt 3rze ( 466 ) liYplCnenFkktfkrilhi 2rh1 ( 324 ) liYc-rspdfriAfqellcl 2vt4A ( 341 ) iiYc-rspdfrkAfkrlla 3pblA ( 381 ) viYttfnieFrkAflkilsc 2ydv ( 286 ) fiyAyrireFrqTFrkiirshvlrqqepfkaa 3v2w ( 309 ) iiytltNkemrrafiri 3oduA ( 300 ) ilyaflgakfktsaqhalts 1u19A ( 304 ) viYimmnkqFrnCmvttlccgknplgddeasttVsktetsqvapa 2z73A ( 313 ) miYsvsHpkFreAIsqtfpwvLtccqfddketeddkdaeteipage aaaaa aaaaaaaaaa
-
Internship Project - A fully Open Chemically Searchable ChEMBL
 For a long time now we have been keen to release a full and freely deployable version of the ChEMBL database with compound search capabilities built in. This has been possible in the past, but complicated by commercial licenses associated with either the databases or the chemical cartridges. There are now a number mature Open Source chemical toolkits available, such as the excellent CDK, and RDKit.
For a long time now we have been keen to release a full and freely deployable version of the ChEMBL database with compound search capabilities built in. This has been possible in the past, but complicated by commercial licenses associated with either the databases or the chemical cartridges. There are now a number mature Open Source chemical toolkits available, such as the excellent CDK, and RDKit.
So with that brief bit of background there is now an opportunity for an intern to work in the ChEMBL group on the project for 2-3 months. The idea is will be to setup a process which:
- Creates a PostgreSQL version of the ChEMBL database (database required by RDKit).
- Install the RDKit chemical cartridge.
- Migrate this setup to Amazon Web Service public image.
- Migrate existing (or new) ChEMBL interface to run off new database and package this up into AWS image.
- Develop scripts to allow new releases of ChEMBL to be processed and uploaded as a new AWS image.
If you are looking for internship this year and have interest in the area of cheminformatics tools and some relevant experience please get in touch (as potential interns, we appreciate you may not have years of industry experience, but we would require you to have previous experience with relational databases and be competent in at least one programming language). Mail us!